June 1, 2016 – Misunderstood maybe, but no longer junk, of the 3 billion letters that spell out the human genome, 98% have remained a mystery. A mere 2% code for proteins, the messengers that help execute the programs that drive us. Because we didn’t understand the remaining 98% we referred to it as junk DNA. Well that’s changing as researchers unlock what the rest of our DNA molecule is all about.
The latest revelation comes from University of Toronto where Professor Benjamin Blencowe and his team at the Donnelly Centre have created LIGR-seq, a tool that interprets DNA messages not specifically coded for proteins. The results of the research is published in the May 19, 2016 edition of Molecular Cell in an article called “Global Mapping of Human RNA-RNA Interactions.”
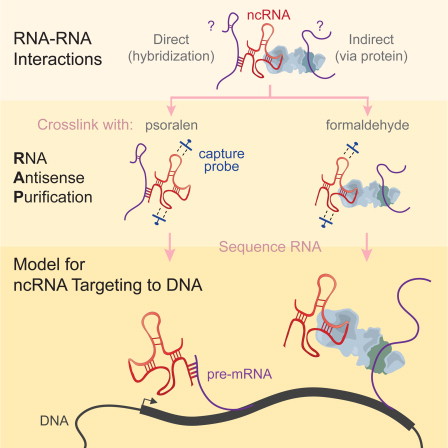
But this article refers to RNA, not DNA, so what does it have to do with the latter? Whereas DNA contains the instructions to make us, RNA molecules serve as a mail service delivering DNA instructions. We’ve understood for sometime that when an RNA envelope gets to its destination and is opened it releases instructions within a cell to generates proteins.
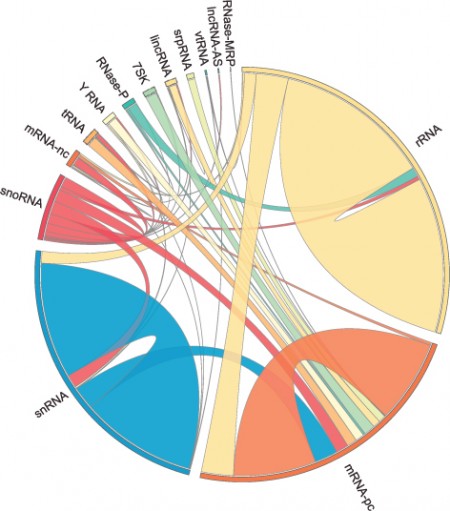
The University of Toronto team’s tool, LIGR-seq, captures the interactions between RNA molecules that are alike and analyzes them. This is giving scientists a better understanding of RNA interactions in living cells. And we are only beginning to understand the many flavours of RNA and what each does with tools like this. In the image below we can see just how many different versions of RNA exist, all detected by LIGR-seq.
For example the researchers have found RNA pairs that influence not what proteins get made but also the quantity. The team is focusing on RNA’s function in the formation of neurons in the context of disease.
The implications of the discovery are well described by Eesha Sharma, a PhD canadidate working in Professor Blencowe’s team. “Up until now, with existing methods, you had to know what you are looking for because they all require you to have some information about the RNA of interest. The power of our method is that you don’t need to preselect your candidates; you can see what’s occurring globally in cells, and use that information to look at interesting things we have not seen before and how they are affecting biology,” states Sharma.